One way to manage digital information is to classify it into a series of categories or a heirarchical taxonomy, and traditionally this was done manually by analysts, who would examine each new document and decide where it should fit. Building and maintaining taxonomies can also be labour intensive, as these will change over time (for a simple example, just consider how political parties change and divide, with factions appearing and disappearing). Search engine technology can be used to automate this classification process and the taxonomy information used as metadata, so that search results can be easily filtered by category, or automatically delivered to those interested in a particular area of the heirarchy.
We’ve been working on an internal project to create a simple taxonomy manager, which we’re releasing today in a pre-alpha state as open source software. Clade lets you import, create and edit taxonomies in a browser-based interface and can then automatically classify a set of documents into the heirarchy you have defined, based on their content. Each taxonomy node is defined by a set of keywords, and the system can also suggest further keywords from documents attached to each node.
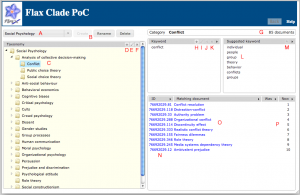
This screenshot shows the main Clade UI, with the controls:
A – dropdown to select a taxonomy
B – buttons to create, rename or delete a taxonomy
C – the main taxonomy tree display
D – button to add a category
E – button to rename a category
F – button to delete a category
G – information about the selected category
H – button to add a category keyword
I – button to edit a keyword
J – button to toggle the sense of a keyword
K – button to delete a keyword
L – suggested keywords
M – button to add a suggested keyword
N – list of matching document IDs
O – list of matching document titles
P – before and after document ranks
Clade is based on Apache Solr and the Stanford Natural Language Processing tools, and is written in Python and Java. You can run it on on either Unix/Linux or Windows platforms – do try it and let us know what you think, we’re very interested in any feedback especially from those who work with and manage taxonomies. The README file details how to install and download it.















Still lots of innovation left in Cambridge despite the loss of Autonomy! Congratulations on the release of this very welcome application. Any thoughts about whether this could be used to manage the development of Managed Metadata schemas in SharePoint?
We haven’t yet implemented a way of exporting the classifications, and importing is currently limited to CSV – but assuming one could work out a suitable integration method with Sharepoint there’s no reason why not.
Great stuff, Charlie. If it’s not too leading a question, how would you position this relative to current proprietary offerings (i.e. any obvious gaps or advances over them)?
Pingback: Reblogging: The WebGenre Blog: The power of genre applied to digital information. By Marina Santini
@Tony – we haven’t carried out any comparative analysis with commercial products, however I can say that an earlier version of this tool was evaluated by a third party and stacked up very well against a commercial alternative in terms of the autoclassification performance, especially given that the commercial alternative is a far more mature product. Also, we couldn’t find many open source taxonomy tools and the ones we did find were rather overcomplicated, which is part of the reason we’ve released Clade.
Hi Charlie, is Clade available on an open test platform too, so we can get an initial idea of how it works with no need to install the server ?
Hi Lila, not currently, but we’ll see what we can do.
Hi Charlie,
We briefly touched on this area during our discussion yesterday. Would it be possible to see a demonstration at the next meeting? I’d also be interested to know if any interest had been shown in using this approach for extracting embedded image/video metadata for autoclassification. We are due to implement a media library in September which extracts EXIF/IPTC/XMP data to a Sharepoint library, and also allows us to merge new/amend metadata to a media derivative.
Regards,
Alan
Hi Alan, of course we’re happy to demo Clade. Regarding extraction of EXIF data, we’ve used the ImageMagick library to do this in the past, and once it’s extracted it could be used like any other metadata for classification, tagging, faceted search etc.
Pingback: progression
Clade is a nice tool on top of Solr. I was successful in getting it working with Solr 4.4 by:
1) changing the solr_url in the settings.py to conform to the multicore standard (e.g.: solr_url = ‘http://localhost:8983/solr/collection1/’ for the default example install of solr)
2) similarly, when copying the conf files over, you have to make sure that you put them into the proper cores fodler… (e.g. solr/collection1/conf NOT solr/conf)
3) replacing the included sunburnt with the latest sunburnt (just named the included sunburnt folder sunburntOLD and copied the new sunburnt folder in) I did this just for good measure as the SolrResponses have changed and I wanted to make sure that the sunburnt I was using to interface with it would handle it properly
4) in lib/taxonomy.py the sunburnt query in get_docs_for_category is being executed with the fields limited to just score=true… this causes a KeyError as doc_id and title are not returned. There is a note in the code to “FIX” it… this may have worked against the old versions of Solr and sunburnt as perhaps that limiting was not working properly in those versions, however, now you need to change the line to: results = query.field_limit([“doc_id”, “title”], score=True).paginate(rows=10).execute()
That brings back all three expected fields to create the desired tuple for return.
I am new to python, Clade, sunburnt and Solr so if anything I have stated here is wrong, please do not hesitate to correct me… just hoping to spare others some pain.
oh, I also Updated the schema config so that the uniqueKey definition for doc_id included the multiValued=”false” attribute
Nice one Alexis! We’ve been hoping to develop Clade further at some point (perhaps by expanding the way classification is done to encompass more complicated expressions for each node) but we haven’t got around to it yet. You’re right about the dupes if you reclassify the same data, this would be a useful improvement.
After several hours with the Stanford classifier, I have found that building distinct language models for each classification and then using log2 ngram probabilities was 3-4x more accurate then standard maxent used in Stanford’s system.
Does the document content need to be stored locally or could use use it to classify web resources (i.e. it fetches the content and analyses it) ?
Currently Clade classifies local documents – but with the addition of a crawler it could easily classify documents gathered from the Web. There are plenty of open source web crawlers out there – for targeted use we’re fans of Scrapy.
Hi,
Really nice tool, many thanks.
Is there a way to set the number of responsive docs to be more than 10? I noticed that there a re a number of “FIX” comments in the taxonomy.py file where “10” seems to be hard coded.
Ideally I’d like to be able to see all responsive docs if possible.
Any help to achieve this would be appreciated.
I am not a python developer.
Hi Mark,
If you change the number set for ‘rows’ in taxonomy.py line 331:
results = query.field_limit(score=True).paginate(rows=10).execute() #FIXME: configurable rowsand re-run the system you should get more documents shown. Bear in mind this may have unintended consequences! We’re working on a new release of Clade with a few improvements, watch this space…
Charlie
Charlie,
Thanks for the response. It worked very well upto about 200 results.
Great to hear that you are working on a new release. I will check back frequently. In our case, our source docs are PDFs. I had to extract the text layer before I could index them. Having a module that could pick up a list of PDF files would be a great addition.
Thanks again
Mark
Glad to hear it. Yes, it would be perfectly feasible to index PDFs, we’d use Apache Tika I suspect.
All:
I got an exception when I run $ ./server.sh $ in the stanford-ner-2011-09-14 directory:
s952275:stanford-ner-2011-09-14 pzhang$ ./server.sh &
[1] 15115
s952275:stanford-ner-2011-09-14 pzhang$ Loading classifier from /Users/pzhang/clade-master/stanford-ner-2011-09-14/classifiers/all.3class.distsim.crf.ser.gz … Exception in thread “main” java.io.FileNotFoundException: classifiers/all.3class.distsim.crf.ser.gz (No such file or directory)
at java.io.FileInputStream.open(Native Method)
at java.io.FileInputStream.(FileInputStream.java:138)
at edu.stanford.nlp.ie.AbstractSequenceClassifier.loadClassifier(AbstractSequenceClassifier.java:1362)
at edu.stanford.nlp.ie.AbstractSequenceClassifier.loadClassifier(AbstractSequenceClassifier.java:1309)
at edu.stanford.nlp.ie.crf.CRFClassifier.getClassifier(CRFClassifier.java:2296)
at edu.stanford.nlp.ie.NERServer.main(NERServer.java:324)
[1]+ Exit 1 ./server.sh
Any help is great.
Pengchu
I downloaded from Stanford nlp site for this version and replaced the current one. Seems work well.
Cannot get it to work 🙁
Got the following error:
Loading classifier from /home/dikchant/applications/clade-master/stanford-ner-2011-09-14/classifiers/all.3class.distsim.crf.ser.gz … Exception in thread “main” java.io.FileNotFoundException: classifiers/all.3class.distsim.crf.ser.gz (No such file or directory)
at java.io.FileInputStream.open(Native Method)
at java.io.FileInputStream.(FileInputStream.java:146)
On download as suggested by Pengchu, I get the following error:
dikchant@dikchant-Latitude-E5440:~/applications/clade-master/stanford-ner-2011-09-14$ ./server.sh
Loading classifier from /home/dikchant/applications/clade-master/stanford-ner-2011-09-14/classifiers/english.all.3class.distsim.crf.ser.gz … Exception in thread “main” java.lang.ClassCastException: java.util.ArrayList cannot be cast to [Ledu.stanford.nlp.util.Index;
at edu.stanford.nlp.ie.crf.CRFClassifier.loadClassifier(CRFClassifier.java:2164)
at edu.stanford.nlp.ie.AbstractSequenceClassifier.loadClassifier(AbstractSequenceClassifier.java:1249)
at edu.stanford.nlp.ie.AbstractSequenceClassifier.loadClassifier(AbstractSequenceClassifier.java:1366)
at edu.stanford.nlp.ie.AbstractSequenceClassifier.loadClassifier(AbstractSequenceClassifier.java:1309)
at edu.stanford.nlp.ie.crf.CRFClassifier.getClassifier(CRFClassifier.java:2296)
at edu.stanford.nlp.ie.NERServer.main(NERServer.java:324)
Try the latest Stanford NLP – as other posters have commented that seems to fix the issue.
i have to extract Person name, location and Organization name using Stanford NER. but i am getting same error
my code is
private CRFClassifier classifier;
private String file =”english.all.3class.distsim.crf.ser.gz”;
CRFClassifier classifier = CRFClassifier.getClassifier(file);
and i am getting error on this line
CRFClassifier classifier = CRFClassifier.getClassifier(file);
and error is
ClassCastException: java.util.Arraylist cannot be cast to [Ledu.stanford.nlp.util.index
someone please me.
Hi Kanza, I think you’d be best asking the Stanford team as we’re not the authors of this software.
Hai…
I am interested to understand clade master tool. I am doing project on taxonomy classification and I have followed all the procedure which you have mentioned in https://github.com/flaxsearch/clade
But I am not understanding how Stanford NER package is used in clade. I went though clasfy.py. I’m very particular in understanding how Stanford NER has been used in taxonomy classification. When I went through documentation of Stanford NER and installed package basically it classify the sentence into location, person, organisation. how this information are used in taxonomy classification.
Kindly let me know the same.